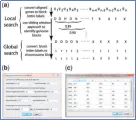
KGBassembler is a tool used to assemble full chromosomes from scaffolds if a karyotype is provided. The KGBassembler is featured with a user-friendly graphical user interface (GUI), allowing users to use automatic assembling of chromosomes based on CCP-based karyotypes and/or to manually edit the layouts of contigs according to in silico generated karyotypes. KGBassembler runs in three steps (Phases I–III). Phase I (Contigs2Blocks) is to identify the 24 blocks in contigs via analyzing BLAT alignments using two algorithms: local and global search. KGBassembler was implemented in C++ with a GUI built with the open-source QT toolbox.
KGBassembler was developed by Chuang Ma in 2011